AnkIdentification
Do you know Anki (https://apps.ankiweb.net/)? This app allows you to create cards to powerfully learn. This program creates a file you can import in Anki to train your plant identification skills. On each card, you'll have plenty of pictures of flowers, leaves, fruits and barks of a plant and then its scientific, vernacular and family names. You can modifiy several parameters and use different ways to import your plant list.
A project made in collaboration with Otyugh.
HOW TO USE
1 Run the code
1- Download main.py, _dict_species.json and _dict_species_by_rarety.json.
2- Download also two large files:
- multimedia.txt (compressed file): https://univangersfr-my.sharepoint.com/:u:/g/personal/aurelien_valentin_etud_univ-angers_fr/EezOrVCe_JpOtDsdrIcK6psBGwAoiV4gtnELEEggryiOQw?e=eIMDJg
- _dict_classification.json: https://univangersfr-my.sharepoint.com/:u:/g/personal/aurelien_valentin_etud_univ-angers_fr/EUk27BFQRKdJlSPFi30-BScBLsY3Kno4XaubfzcaCljVYg?e=5fcA7V
3- While the files are downloading, you'll have to choose the species you want to learn. Three ways are proposed:
- You can use the default list of all species found in France (sorted by their frequence in this country). Its generation is included in main.py.
- You can go on GeoPl@ntNet (https://identify.plantnet.org/prediction) and select an area. An AI will list you the 100 most common plants in there. Keep the "sorting by GBIF" and download the CSV file. You'll have to specify the name of your file in the SPECIE_LIST_PATH_IF_GBIF_METHOD variable in main.py. France_GBIF.csv is an example of file you may obtain.
- You can create your own list of plants you want to learn. In this case, please create a txt file with one specie by line. You'll have to specify the name of your file in the MY_OWN_LIST_PATH variable in main.py. You can take Species_list.txt as a template.
4- Change parameters of main.py. Everything is written in the comments.
5- Install the genanki module by writting "pip install genanki" in your terminal. It allows you to create Anki decks and cards with Python, further information here: https://github.com/kerrickstaley/genanki.
6- If all the files are downloaded, put them in the same directory and then run the code to obtain AnkIdentification.apkg. The output with the 200 most common plants according to the default list is given.
2 Use Anki
If you don't know Anki, here are the steps to use your output file.
1- Download Anki here: https://apps.ankiweb.net/.
2- Create your account, you'll be able to find your cards anywhere with the web application (https://ankiweb.net/), the android version (https://play.google.com/store/apps/details?id=com.ichi2.anki) and/or the apple one which is not free (https://apps.apple.com/us/app/ankimobile-flashcards/id373493387).
3- At the bottom of the app, you'll find the import button to add the deck previously generated (AnkIdentification.apkg).
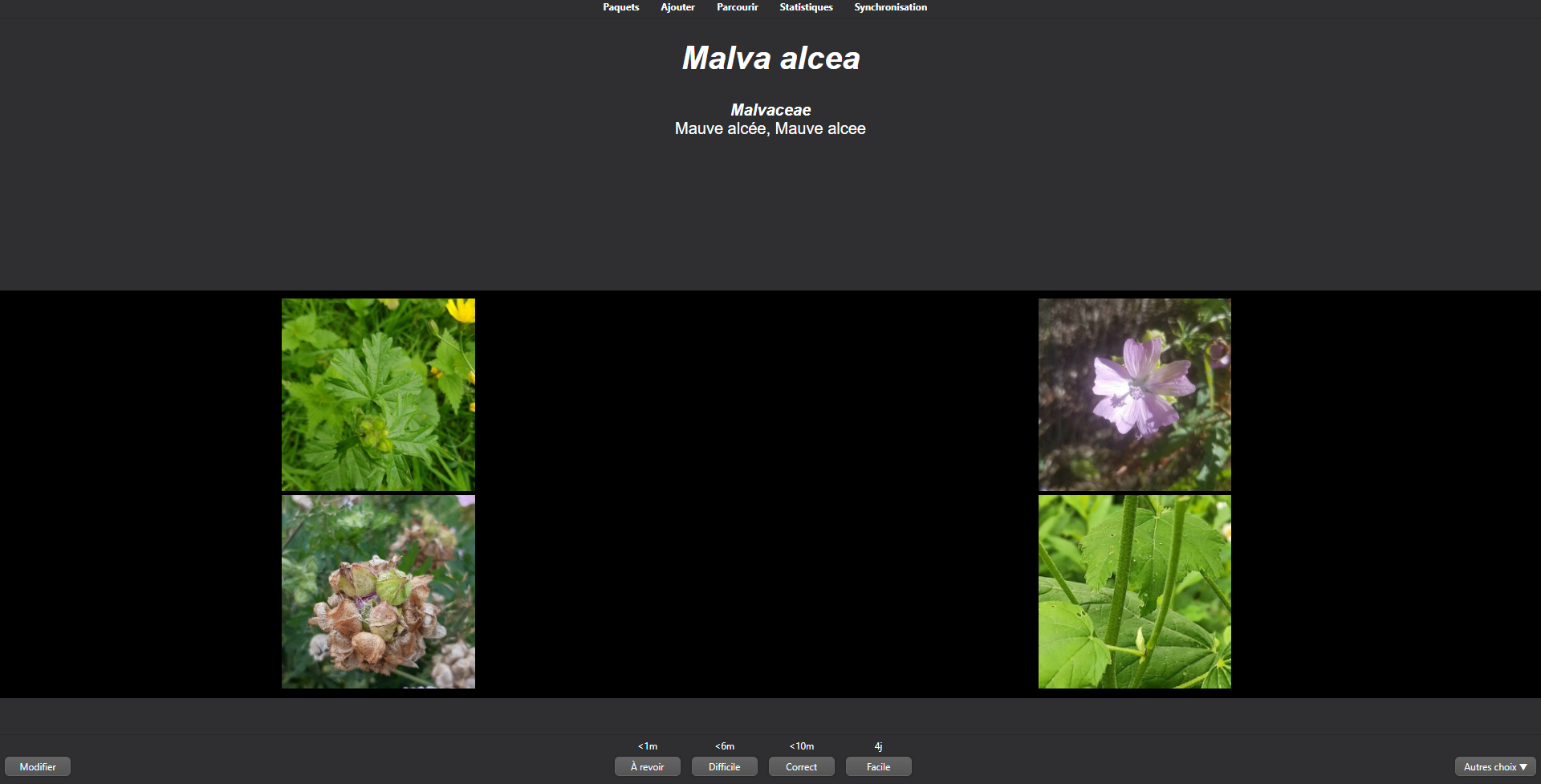
4- You can then click on "AnkIdentification" and "Study now" and see four images: leaves on the top left, flowers on the top right, fruits and the bottom left and barks on the bottom right.
- A left click will change an image for another
- A right click will load an image in full screen
- A long click will return to the previous image
- Let your cursor on an image to see the author of it
- On the top left you can change the resolution of the pictures depending on your internet connection
5- Press space or click "Show the answer" to have the scientific name of the plant, its family and some vernacular names.
6- Regarding how you identified well or not the species, you can decide to review this card rapidly or not. You'll then see difficult card more often than the easy one.
7- To browse cards, press "B". You can search different species and find tags on the left panel to order plants by family.
8- Get more info about Anki on the doc (https://docs.ankiweb.net/) or in this video for example: https://youtu.be/vm4oooKANzU.
NOTA BENE
- Supplementary files are available to see how dictionaries from raw datasets have been obtained for the main code.
- Prefer to use Anki on computer because the cards don't suit well with smartphones (right click unpossible, response time different...)
- Scientific names and family change all the time... The data of this project have been downloaded in 2022 and are derived from the APG IV (Angiosperm Phylogeny Group). They may be outdated in the years to come...
- Some species only get an English vernacular name. For these plants, French users will find an "en" before the only name found.
- Just in case, in the link fields you won't find whole URLs, add "https://bs.plantnet.org/image/o/" before an element to get the original picture link.
SOURCES
Here are the references to the original files used in this project:
- multimedia.txt (for the links) and 0229588-220831081235567.csv (for the list of species where links are available) are from Pl@ntNet: https://www.gbif.org/dataset/7a3679ef-5582-4aaa-81f0-8c2545cafc81
- classification.txt (to associate a family to each species) is downloaded from the World Flora Online (more up-to-date than Pl@ntNet): http://www.worldfloraonline.org/downloadData
- specieswiki-20221201-pages-meta-current.xml used to produce _dict_species.json (to attribute common names) is from the whole Wikispecies pages (https://dumps.wikimedia.org/specieswiki/20221201/). Make a list of all vernacular names is not a major purpose of Wikispecies so many of them may be missing...
Don't hesitate to comment this work, which is my first to be published. So I thank you in advance ^^!